Working with DataFrames
Contents
Working with DataFrames¶
Now we will go to a territory which might be somewhat familiar if you have previously used R (or Matlab). We will import some packages, read the contents of a file and store it in a data-frame.
Importing packages¶
One of the great things about python that very often you don ‘t have to implement something yourself. It may be already implemented in the many freely accessible python packages, often referred to as the python ecosystem.
If you are working on your local computer, you can install python packages e.g. with the tools pip or conda.
Inside a notebook, hosted on the cloud (e.g. Google Colab), you can install packages via the following code.
!pip install pandas nilearn
Requirement already satisfied: pandas in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (1.3.2)
Requirement already satisfied: nilearn in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (0.8.0)
Requirement already satisfied: python-dateutil>=2.7.3 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from pandas) (2.8.2)
Requirement already satisfied: pytz>=2017.3 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from pandas) (2021.1)
Requirement already satisfied: numpy>=1.17.3 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from pandas) (1.19.5)
Requirement already satisfied: joblib>=0.12 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from nilearn) (1.0.1)
Requirement already satisfied: scipy>=1.2 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from nilearn) (1.7.1)
Requirement already satisfied: requests>=2 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from nilearn) (2.26.0)
Requirement already satisfied: nibabel>=2.5 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from nilearn) (3.2.1)
Requirement already satisfied: scikit-learn>=0.21 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from nilearn) (0.24.2)
Requirement already satisfied: packaging>=14.3 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from nibabel>=2.5->nilearn) (21.0)
Requirement already satisfied: pyparsing>=2.0.2 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from packaging>=14.3->nibabel>=2.5->nilearn) (2.4.7)
Requirement already satisfied: six>=1.5 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from python-dateutil>=2.7.3->pandas) (1.16.0)
Requirement already satisfied: certifi>=2017.4.17 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from requests>=2->nilearn) (2021.5.30)
Requirement already satisfied: charset-normalizer~=2.0.0 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from requests>=2->nilearn) (2.0.4)
Requirement already satisfied: idna<4,>=2.5 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from requests>=2->nilearn) (3.2)
Requirement already satisfied: urllib3<1.27,>=1.21.1 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from requests>=2->nilearn) (1.26.6)
Requirement already satisfied: threadpoolctl>=2.0.0 in /home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages (from scikit-learn>=0.21->nilearn) (2.2.0)
WARNING: You are using pip version 21.2.4; however, version 22.0.3 is available.
You should consider upgrading via the '/home/tspisak/src/RPN-signature/venv/bin/python -m pip install --upgrade pip' command.
This command installed two packages needed for the rest of this notebook: pandas and nilearn.
Once a package is installed on the system, the only thing we must do is to import the packages, in order to tell python that we intend to use it in your code.
Now, we will import the package called pandas, a package that provides powerful R-like dataframes to store and manipulate your data. For convenience, we also specify that from now on, we would like to refer to pandas as pd, for short.
import pandas as pd
Loading some data¶
Now let’s load some example data. Throughout the book, we will use brain cortical volume data obtained from the publicly available “Information eXtraction from Images” (IXI) dataset
See also
See supplement X for how raw IXI anatomical MRI data was processed with Freesurfer. For more information on analyzing anatomical MR images with Freesurfer e.g. on Andy’s blog.
df=pd.read_csv("https://raw.githubusercontent.com/pni-lab/predmod_lecture/master/ex_data/IXI/ixi.csv")
df
| ID | Age | lh_bankssts_volume | lh_caudalanteriorcingulate_volume | lh_caudalmiddlefrontal_volume | lh_cuneus_volume | lh_entorhinal_volume | lh_fusiform_volume | lh_inferiorparietal_volume | lh_inferiortemporal_volume | ... | rh_rostralanteriorcingulate_volume | rh_rostralmiddlefrontal_volume | rh_superiorfrontal_volume | rh_superiorparietal_volume | rh_superiortemporal_volume | rh_supramarginal_volume | rh_frontalpole_volume | rh_temporalpole_volume | rh_transversetemporal_volume | rh_insula_volume | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 35.800137 | 2188 | 2368 | 6562 | 2459 | 1561 | 9281 | 14136 | 10797 | ... | 2555 | 17309 | 21210 | 12291 | 12287 | 10848 | 1033 | 2269 | 1170 | 6915 |
| 1 | 12 | 38.781656 | 2717 | 2626 | 6621 | 3170 | 2835 | 8870 | 14813 | 11961 | ... | 3260 | 19044 | 24651 | 13871 | 13948 | 11975 | 1100 | 2865 | 1167 | 6941 |
| 2 | 13 | 46.710472 | 2101 | 2488 | 5437 | 2347 | 1859 | 9200 | 16900 | 11675 | ... | 2682 | 17653 | 23804 | 10977 | 12931 | 15127 | 975 | 2099 | 1032 | 7395 |
| 3 | 14 | 34.236824 | 1925 | 1983 | 5153 | 2497 | 2207 | 7686 | 12786 | 8433 | ... | 2120 | 15070 | 21001 | 10993 | 10890 | 10453 | 891 | 2122 | 958 | 6063 |
| 4 | 15 | 24.284736 | 2535 | 1802 | 5461 | 2496 | 1875 | 6859 | 14187 | 7897 | ... | 2825 | 13027 | 21865 | 10651 | 12686 | 11400 | 1185 | 2207 | 1344 | 8218 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 633 | 648 | 47.723477 | 2009 | 2270 | 5439 | 2684 | 1729 | 9029 | 13407 | 10971 | ... | 3236 | 15464 | 21427 | 11524 | 12869 | 11373 | 814 | 2419 | 1360 | 7423 |
| 634 | 651 | 50.395619 | 1776 | 1532 | 5797 | 2954 | 1992 | 9131 | 13201 | 9005 | ... | 2146 | 11883 | 18137 | 13668 | 10833 | 8420 | 751 | 2304 | 1010 | 6032 |
| 635 | 652 | 42.989733 | 2315 | 1898 | 6234 | 3285 | 1857 | 10453 | 15412 | 10639 | ... | 2931 | 16905 | 25113 | 13148 | 14489 | 13440 | 731 | 2725 | 1525 | 7481 |
| 636 | 653 | 46.220397 | 2411 | 2033 | 5808 | 4012 | 2854 | 9723 | 16169 | 12070 | ... | 2637 | 15636 | 19449 | 13207 | 13480 | 11459 | 1049 | 2855 | 1078 | 7186 |
| 637 | 662 | 41.741273 | 2482 | 1923 | 4686 | 3738 | 1910 | 10996 | 16778 | 10390 | ... | 3492 | 16781 | 20968 | 13379 | 15787 | 12850 | 1069 | 2521 | 1062 | 7931 |
638 rows × 70 columns
The first two columns of our example dataframe are participant ID and age, then we have a total of 68 cortical volume values, each computed in 34 regions. Rows correspond to participants: a total of N=638. The brain regions were delineated by the as based on the Destrieux brain atlas [5], shown below.
from nilearn import datasets, plotting
destrieux_atlas = datasets.fetch_atlas_surf_destrieux(verbose=0)
fsaverage = datasets.fetch_surf_fsaverage()
plotting.plot_surf_roi(fsaverage['pial_left'], roi_map=destrieux_atlas['map_left'],
hemi='left', view='lateral',
bg_map=fsaverage['sulc_left'], bg_on_data=True)
plotting.show()
/home/tspisak/src/RPN-signature/venv/lib/python3.8/site-packages/nilearn/datasets/__init__.py:86: FutureWarning: Fetchers from the nilearn.datasets module will be updated in version 0.9 to return python strings instead of bytes and Pandas dataframes instead of Numpy arrays.
warn("Fetchers from the nilearn.datasets module will be "
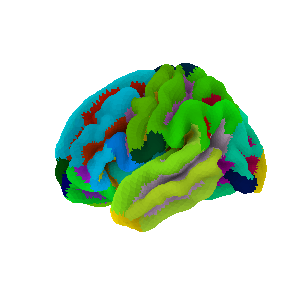
Tip
Click on “Click to show” to reveal the python code used for visualizing the atlas. Visualization was done with the python package ‘nilearn’.
See also
Nilearn is a very powerful package for machin elearning with neuroimaging. Check out the nilearn example gallery to have a better impression.
Dataframe slicing¶
You can get one or multiple columns from a dataframe by slicing it. Let’s extract the age of the participants.
df['Age']
0 35.800137
1 38.781656
2 46.710472
3 34.236824
4 24.284736
...
633 47.723477
634 50.395619
635 42.989733
636 46.220397
637 41.741273
Name: Age, Length: 638, dtype: float64
Now let’s slice the data frame to obtain ID, Age and the volume of the brain region called right superior frontal cortex and obtain the first 5 participants only. To do so you must slice with a list of column names, that’s whz we have double square brackets (outer: slicing, inner: list). Then we call head, to get the ‘head’ of the table.
df_superiorfrontal = df[['ID', 'Age', 'rh_superiorfrontal_volume']]
df_superiorfrontal.head(5)
| ID | Age | rh_superiorfrontal_volume | |
|---|---|---|---|
| 0 | 2 | 35.800137 | 21210 |
| 1 | 12 | 38.781656 | 24651 |
| 2 | 13 | 46.710472 | 23804 |
| 3 | 14 | 34.236824 | 21001 |
| 4 | 15 | 24.284736 | 21865 |
It is very easy to filter you data frame by the values of one of the columns.
df_superiorfrontal[df_superiorfrontal['Age']>30]
| ID | Age | rh_superiorfrontal_volume | |
|---|---|---|---|
| 0 | 2 | 35.800137 | 21210 |
| 1 | 12 | 38.781656 | 24651 |
| 2 | 13 | 46.710472 | 23804 |
| 3 | 14 | 34.236824 | 21001 |
| 5 | 16 | 55.167693 | 20534 |
| ... | ... | ... | ... |
| 633 | 648 | 47.723477 | 21427 |
| 634 | 651 | 50.395619 | 18137 |
| 635 | 652 | 42.989733 | 25113 |
| 636 | 653 | 46.220397 | 19449 |
| 637 | 662 | 41.741273 | 20968 |
534 rows × 3 columns
Plotting¶
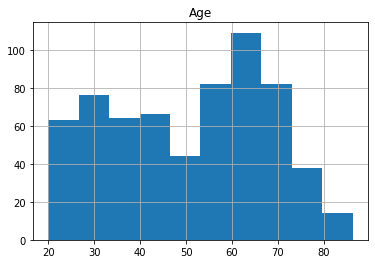
Pandas dataframes provide build-in plotting functions. Let’s see how the age of the participants is distributed.
df.hist('Age')
array([[<AxesSubplot:title={'center':'Age'}>]], dtype=object)
Now, let’s plot the volume of our previously selected brain region against age.
df.plot.scatter(x='Age', y='rh_superiorfrontal_volume')
<AxesSubplot:xlabel='Age', ylabel='rh_superiorfrontal_volume'>
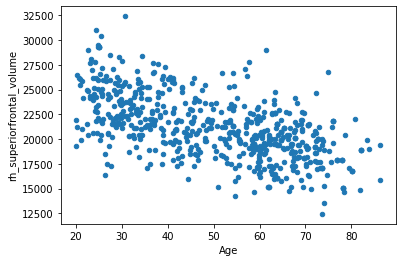
Looks like an inverse association… In the next section, we will see how we can take advantage of this single association in order to predict the age of a participant. And also how we can get into trouble when we want to improve the prediction by adding more regions.
See also
Pandas has an excellent leighweight “Getting Started” tutorial.
Excercise 1.2
Launch this notebook interactively in Google Colab (by clicking the rocket) and modify the code to plot out other variables, e.g. rh_superiorfrontal_volume vs. lh_superiorfrontal_volume.
Excercise 1.3
Launch this notebook interactively in Google Colab (by clicking the rocket) and modify it to load in your own dataset. You can directly upload your dataset file in colab (Select “Files” on the left) or you can also mount your google drive account.
Is you dataframe as Excel spreadsheet? No problem! Check out the function read_excel in pandas.
